
Links
2025 & Preprints
Integration of 168,000 samples reveals global patterns of the human gut microbiome
R. J. Abdill, S. P. Graham, M. Ahmadian, P. Hicks, A. Chetty, D. McDonald, P. Ferretti, E. Gibbons, M. Rossi, A. Krishnan, V. Rubinetti, F. W. Albert, C. S. Greene, S. Davis, and R. BlekhmanCell, 2025
Gut microbiome data underscores gaps in global representation. University of Chicago News
The Human Microbiome Compendium on the cover of Cell. Cell
Microbiomap.org - the Human Microbiome Compendium portal microbiomap.org
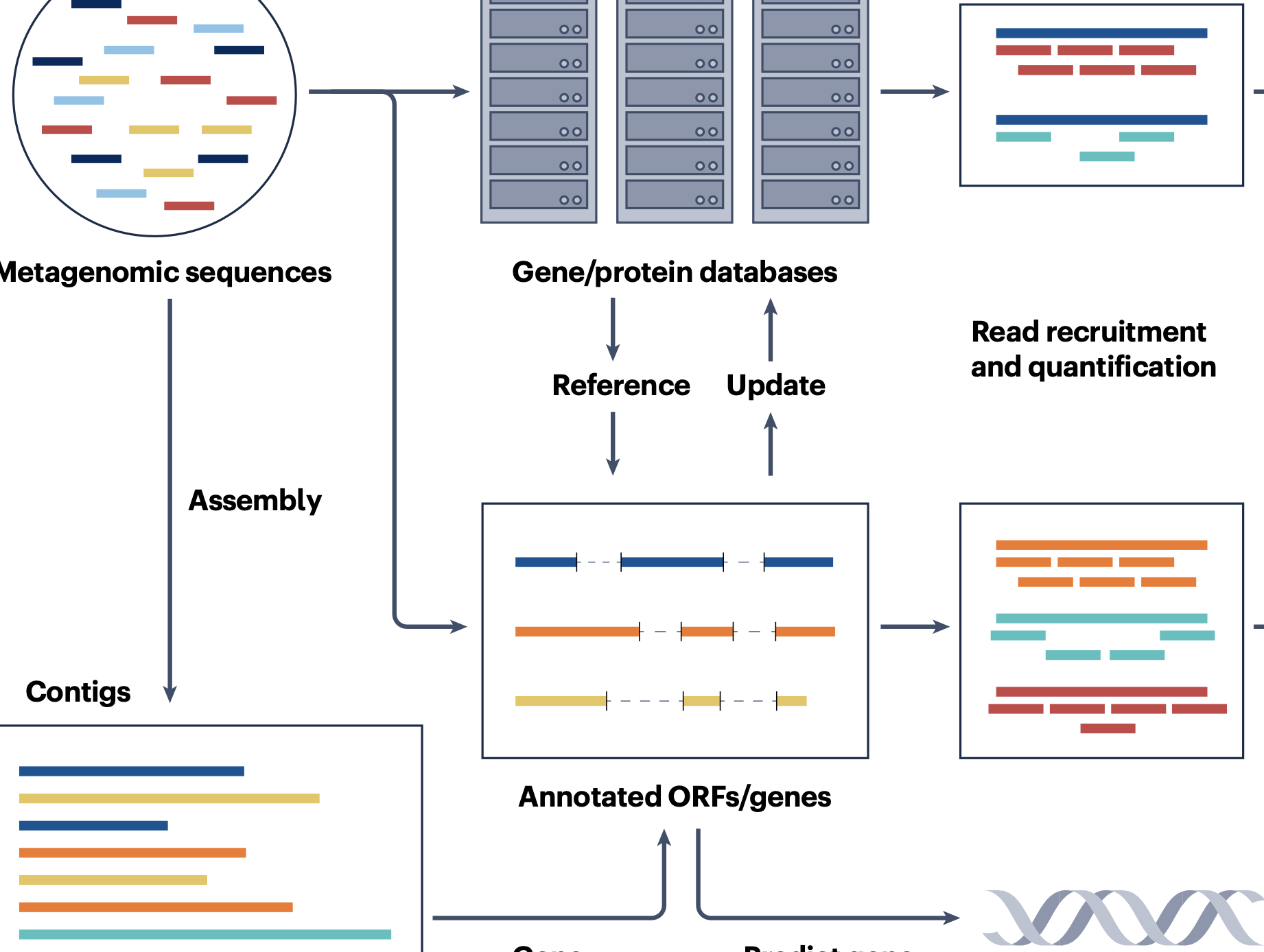
Analysis of metagenomic data
S. Liu, J. Rodriguez, V. Munteanu, C. Ronkowski, N. K. Sharma, M. Alser, F. Andreace, R. Blekhman, D. Błaszczyk, R. Chikhi, K. A. Crandall, K. Della Libera, D. Francis, A. Frolova, A. Shahar Gancz, N. E. Huntley, P. Jaiswal, T. Kosciolek, P. P. Łabaj, W. Łabaj, T. Luan, C. Mason, A. M. Moustafa, H. Muralidharan, O. Mutlu, N. Mansouri Ghiasi, A. Rahnavard, F. Sun, S. Tian, B. T. Tierney, E. Van Syoc, R. Vicedomini, J. P. Zackular, A. Zelikovsky, K. Zielińska, E. Ganda, E. R. Davenport, M. Pop, D. Koslicki, and S. Mangul.Nature Reviews Methods Primers, 2025
Cardiometabolic disease risk in gorillas is associated with altered gut microbial metabolism
S. Davison, A. Mascellani Bergo, Z. Ward, A. Sackett, A. Strykova, J. Diógenes Jaimes, D. Travis, J. B. Clayton, H. W. Murphy, M. D. Danforth, B. K. Smith, R. Blekhman, T. Fuh, F. S. N. Singa, J. Havlik, K. Petrzelkova, and A. Gomez.npj Biofilms and Microbiomes volume , 2025
Assembly, stability, and dynamics of the infant gut microbiome are linked to bacterial strains and functions in mother's milk
M. Allert, P. Ferretti, K. E. Johnson, T. Heisel, S. Gonia, D. Knights, D. A. Fields, F. W. Albert, E. W. Demerath, C. A. Gale, and R. BlekhmanBioRxiv preprint
2024
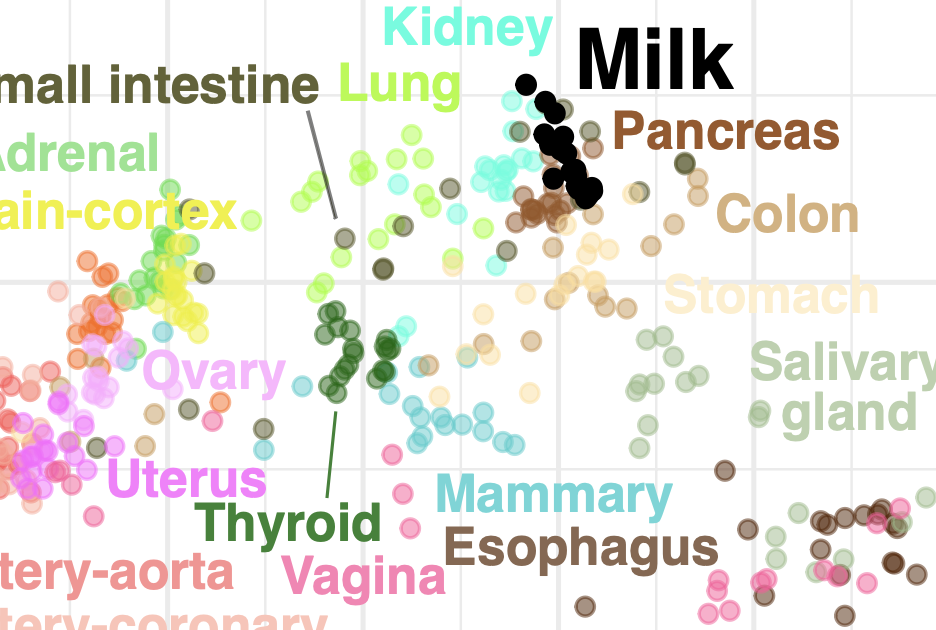
Human milk variation is shaped by maternal genetics and impacts the infant gut microbiome
K. E. Johnson, T. Heisel, M. Allert, A. Fürst, N. Yerabandi, D. Knights, K. M. Jacobs, E. F. Lock, L. Bode, D. A. Fields, M. C. Rudolph, C. A. Gale, F. W. Albert, E. W. Demerath, and R. BlekhmanCell Genomics, 2024
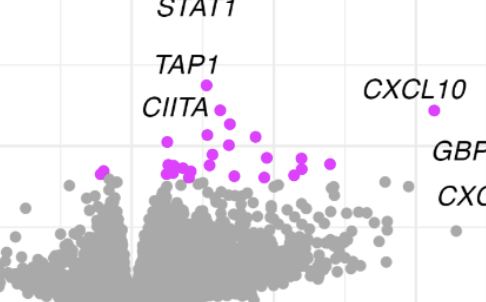
Human Cytomegalovirus in breast milk is associated with milk composition and the infant gut microbiome and growth
K. E. Johnson, N. Hernandez-Alvarado, M. Blackstad. Heisel, M. Allert, D. A. Fields, E. Isganaitis, K. M. Jacobs, D. Knights, E. F. Lock, M. C. Rudolph, C. A. Gale, M. R. Schleiss, F. W. Albert, E. W. Demerath, and R. BlekhmanNature Communications, 2024
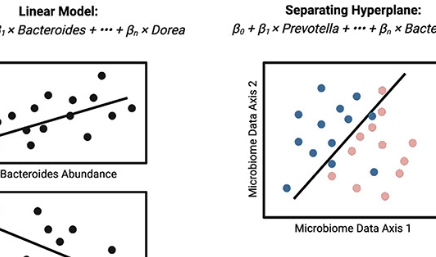
Multi-omic approaches for host-microbiome data integration
A. Chetty and R. BlekhmanGut Microbes, 2024
Social and environmental predictors of gut microbiome age in wild baboons
M. Dasari, K. Roche, D. Jansen, J. Anderson, S. C. Alberts, J. Tung, J. A. Gilbert, R. Blekhman, S. Mukherjee, and E. A. ArchieeLife, 2024
Temporal patterns of gut microbiota in lemurs (Eulemur rubriventer) living in intact and disturbed habitats in a novel sample type
L. Grieneisen, A. Hays, E. Cook, R. Blekhman, and S. TecotAmerican Journal of Primatology
2023
Universal gut microbial relationships in the gut microbiome of wild baboons
K. Roche, J. R. Björk, M. Dasari, L. Grieneisen, D. Jansen, T. J. Gould, L. R. Gesquiere, L. B. Barreiro, S. C. Alberts, R. Blekhman, J. A. Gilbert, J. Tung, S. Mukherjee, and E. A. ArchieeLife, 2023
How longitudinal data can contribute to our understanding of host genetic effects on the gut microbiome
L. Grieneisen, R. Blekhman, and E. A. ArchieGut Microbes, 2023
Microbes with higher metabolic independence are enriched in human gut microbiomes under stress
I. Veseli, Y. T. Chen, M. Schechter, C. Vanni, E. C. Fogarty, A. Watson, B. Jabri, R. Blekhman, A. D. Willis, M. K. Yu, A. Fernandez Guerra, J. Füssel, and A. Murat EreneLife, 2023
2022
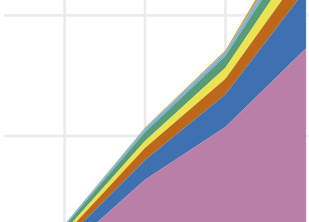
Public human microbiome data are dominated by highly developed countries
R. J. Abdill, E. M. Adamowicz, and R. BlekhmanPLOS Biology, 2022
Studies of human microbiome have ignored the developing world, potentially compromising treatments. Science
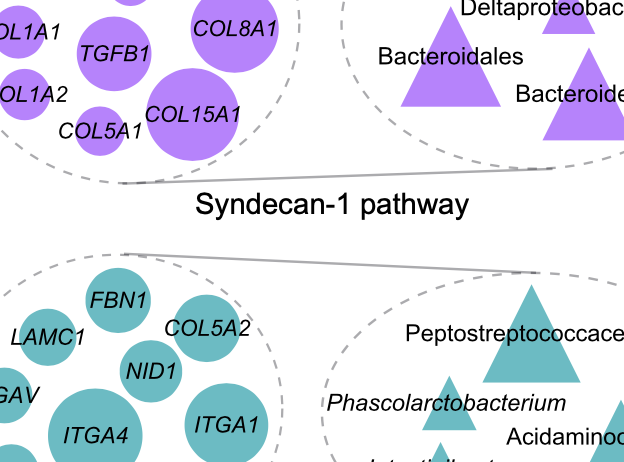
Identification of shared and disease-specific host gene–microbiome associations across human diseases using multi-omic integration
S. Priya, M. B. Burns, T. Ward, R. Mars, E. Adamowicz, E. F. Lock, P. C. Kashyap, D. Knights, and R. BlekhmanNature Microbiology, 2022
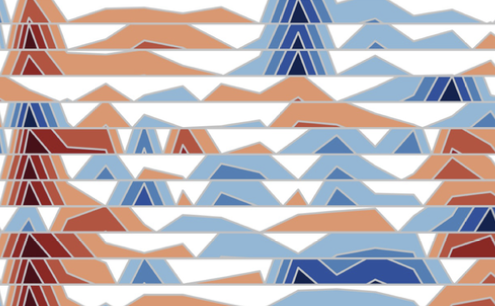
BiomeHorizon: visualizing microbiome time series data in R
I. Fink, R. J. Abdill, R. Blekhman, and L. GrieneisenmSystems, 2022
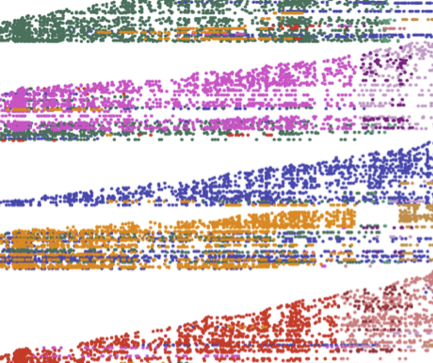
Synchrony and idiosyncrasy in the gut microbiome of wild baboons
J. R. Björk, M. Dasari, L. Grieneisen, T. J. Gould, J.-C. Grenier, V. Yotova, N. Gottel, D. Jansen, K. Roche, S. Mukherjee, L. R. Gesquiere, J. B. Gordon, N. H. Learn, T. L. Wango, R. S. Mututua, J. K. Warutere, L. Siodi, L. B. Barreiro, S. C. Alberts, J. A. Gilbert, J. Tung, R. Blekhman, and E. A. ArchieNature Ecology & Evolution, 2022
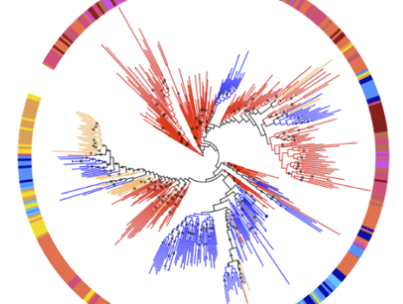
Codiversification of gut microbiota with humans
T. A. Suzuki, L. Fitzstevens, V. T. Schmidt, H. Enav, K. Huus, M. Mbong Ngwese, A. Grieshammer, A. Pfleiderer, B. R. Adegbite, J. F. Zinsou, M. Esen, T. P. Velavan, A. A. Adegnika, L. H. Song, T. D. Spector, A. L. Muehlbauer, N. Marchi, H. Kang, L. Maier, Ran Blekhman, L. Segurel, G. Ko, N. D. Youngblut, P. Kremsner, and R. E. LeyScience, 2022
2021
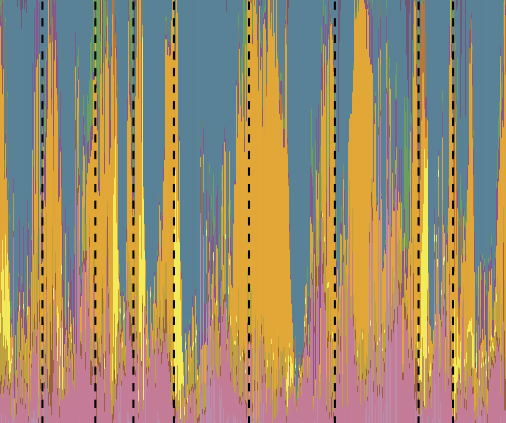
Gut microbiome heritability is nearly universal but environmentally contingent
L. Grieneisen, M. Dasari, T. J. Gould, J. R. Björk, J.-C. Grenier, V. Yotova, D. Jansen, N. Gottel, J. B. Gordon, N. H. Learn, L. R. Gesquiere, T. L. Wango, R. S. Mututua, J. K. Warutere, L. Siodi, J. A. Gilbert, L. B. Barreiro, S. C. Alberts, J. Tung, E. A. Archie, and R. BlekhmanScience, 2021
Host genetics influence the gut microbiome. Science
Gut microbiome affected by genetics more than once thought. GEN
Role of host genetics on gut microbiome is near-universal, but environmentally-dependent. University of Minnesota News
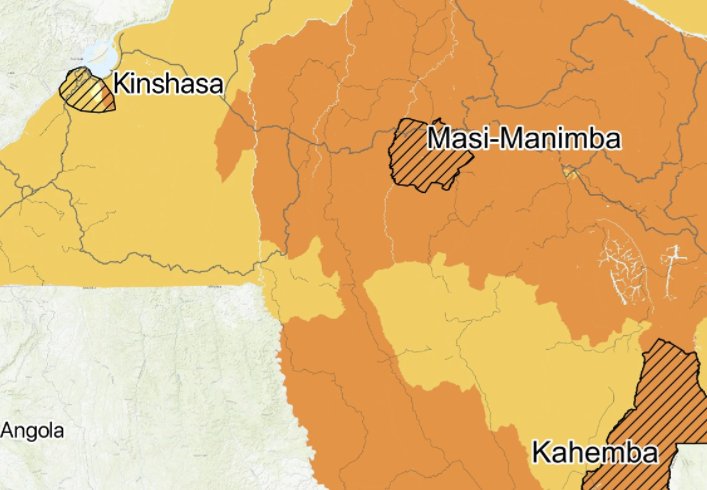
The gut microbiome in konzo
M. S. Bramble, N. Vashist, A. Ko, S. Priya, C. Musasa, A. Mathieu, D. Spencer, M. Kasendue, P. Dilufwasayo, K. Karume, J. Nsibu, H. Manya, M. Uy, B. Colwell, M. Boivin, J. Mayambu, D. Okitundu, A. Droit, D. Mumba Ngoyi, R. Blekhman, D. Tshala-Katumbay, and E. Vilain.Nature Communications, 2021
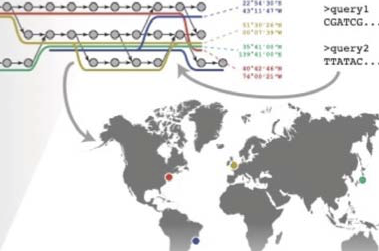
A global metagenomic map of urban microbiomes and antimicrobial resistance
D. Danko and The International MetaSUB ConsortiumCell, 2021
Subway swabbers find a microbe jungle and thousands of new species. New York Times
Cities have their own distinct microbial fingerprints. Science
Interspecies variation in hominid gut microbiota controls host gene regulation
A. L. Muehlbauer, A. L. Richards, A. Alazizi, M. Burns, A. Gomez, J. B. Clayton, K. Petrzelkova, C. Cascardo, J. Resztak, X. Wen, R. Pique-Regi, F. Luca, and R. BlekhmanCell Reports, 2021
Multi-omics analyses show disease, diet, and transcriptome interactions with the virome
K. Mihindukulasuriya, R. A. T. Mars, A. J. Johnson, T. Ward, S. Priya, H. R. Lekatz, K. R. Kalari, L. Droit, T. Zheng, R. Blekhman, M. D’Amato, G. Farrugia, D. Knights, S. A. Handley, and P. C. Kashyap.Gastroenterology, 2021
2020
Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome
R. A. T. Mars, Y. Yang, T. Ward, S. Priya, H. R. Lekatz, X. Tang, Z. Sun, K. R. Kalari, T. Korem, Y. Bhattarai, M. Houtti, A. J. Cole, T. Ordog, M. Grover, M. D’Amato, M. Camilleri, E. Elinav, E. Segal, R. Blekhman, G. Farrugia, J. Swann, D. Knights, and P. C. Kashyap.Cell, 2020
Interactions between the gut microbiome and host gene regulation in cystic fibrosis
G. Dayama, S. Priya, D. E. Niccum, A. Khoruts, and R. BlekhmanGenome Medicine, 2020

International authorship and collaboration across bioRxiv preprints
R. J. Abdill, E. M. Adamowicz, and R. BlekhmaneLife, 2020
Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment
K. R. Chng and The International MetaSUB ConsortiumNature Medicine, 2020
Microbial control of host gene regulation and the evolution of host–microbiome interactions in primates
L. Grieneisen, A. L. Muehlbauer, and R. BlekhmanPhilosophical Transactions of the Royal Society B, 2020
Traditional human populations and nonhuman primates show parallel gut microbiome adaptations to analogous ecological conditions
A. K. Sharma, K. J. Petrželková, B. Pafčo, C. Jost Robinson, T. Fuh, B. A. Wilson, R. M. Stumpf, M. G. Torralba, R. Blekhman, B. White, K. E. Nelson, S. R. Leigh, and A. GomezmSystems, 2020
2019
Genes, geology and germs: gut microbiota across a primate hybrid zone are explained by site soil properties, not host species
L. E. Grieneisen, M. J. E. Charpentier, S. C. Alberts, R. Blekhman, G. Bradburd, J. Tung, and E. A. ArchieProceedings of the Royal Society B, 2019
Baboons’ gut makeup is determined mostly by soil, not genetics. Science

Megaphages infect Prevotella and variants are widespread in gut microbiomes
A. E. Devoto, J. M. Santini, M. R. Olm, K. Anantharaman, P. Munk, J. Tung, E. A. Archie, P. J. Turnbaugh, K. D. Seed, R. Blekhman, F. M. Aarestrup, B. C. Thomas, and J. F. BanfieldNature Microbiology, 2019

Improving the usability and archival stability of bioinformatics software
S. Mangul, L. Martin, E. Eskin, and R. BlekhmanGenome Biology, 2019
Rxivist.org: Sorting biology preprints using social media and readership metrics
R. Abdill and R. BlekhmanPLOS Biology, 2019
Mapping gastrointestinal gene expression patterns in wild primates and humans via fecal RNA-seq
A. K. Sharma, B. Pafčo, K. Vlčková, B. Červená, J. Kreisinger, S. Davison, K. Beeri, T. Fuh, S. R. Leigh, M. B. Burns, R. Blekhman, K. J. Petrželková, and A. GomezBMC Genomics, 2019
Gut microbiota has a widespread and modifiable effect on host gene regulation
A. L. Richards, A. L. Muehlbauer, A. Alazizi, M. B. Burns, A. Findley, F. Messina, T. J. Gould, C. Cascardo, R. Pique-Regi, R. Blekhman, and F. Luca.mSystems, 2019

Challenges and recommendations to improve the installability and archival stability of omics computational tools
S. Mangul, T. Mosqueiro, R. J. Abdill, D. Duong, K. Mitchell, V. Sarwal, B. Hill, J. Brito, R. Littman, B. Statz, A. Lam, G. Dayama, L. Grieneisen, L. Martin, J. Flint, E. Eskin, and R. BlekhmanPLOS Biology, 2019

Plasticity in the human gut microbiome defies evolutionary constraints
A. Gomez, A. Sharma, E. Mallott, K. Petrzelkova, C. Jost Robinson, C. Yeoman, F. Carbonero, B. Pafco, J. Rothman, A. Ulanov, K. Vlckova, K. R. Amato, S. Schnorr, N. Dominy, D. Modry, A. Todd, M. Torralba, K. Nelson, M. Burns, R. Blekhman, M. Remis, R. Stumpf, B. Wilson, H. Gaskins, P. Garber, B. White, and S. Leigh.mSphere, 2019

Population dynamics of the human gut microbiome: change is the only constant
S. Priya and R. BlekhmanGenome Biology, 2019
2018
Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment
M. B. Burns, E. Montassier, J. Abrahante, S. Priya, D. E. Niccum, A. Khoruts, T. K. Starr, D. Knights, and R. BlekhmanPLOS Genetics, 2018

Interaction between host microRNAs and the gut microbiota in colorectal cancer
C. Yuan, M. B. Burns, S. Subramanian, and R. BlekhmanmSystems, 2018
Gut microbiota diversity across ethnicities in the United States
A. W. Brooks, S. Priya, R. Blekhman, and S. R. Bordenstein.PLOS Biology, 2018
Ethnicity proves reliable indicator of what microbes thrive in the gut Research News @Vanderbilt
Integrating tumor genomics into studies of the microbiome in colorectal cancer
M. B. Burns and R. BlekhmanGut Microbes, 2018 (Invited Addendum)
The gut microbiome of nonhuman primates: Lessons in ecology and evolution
J. B. Clayton, A. Gomez, K. Amato, D. Knights, D. A. Travis, R. Blekhman, R. Knight, S. Leigh, R. Stumpf, T. Wolf, K. E. Glander, F. Cabana, and T. J. Johnson.American Journal of Primatology, 2018

Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers
V. L. Hale, P. Jeraldo, J. Chen, M. Mundy, J. Yao, S. Priya, G. Keeney, K. Lyke, J. Ridlon, B. A. White, A. J. French, S. Thibodeau, C. Diener, O. Resendis-Antonio, J. Gransee, T. Dutta, X.-M. T. Petterson, J. Sung, R. Blekhman, L. Boardman, D. Larson, H. Nelson, and N. ChiaGenome Medicine
Transposon mutagenesis screen in mice identifies TM9SF2 as a novel colorectal cancer oncogene.
C. Clark, M. Maile, P. Blaney, S. Hellweg, A. Strauss, W. Durose, S Priya^, M. B. Burns^, S. Subramanian, R. Blekhman, J. Abrahante, and T. StarrScientific Reports
2017
Functional genomics of host–microbiome interactions in humans
F. Luca, S. S. Kupfer, D. Knights, A. Khoruts, and R. BlekhmanTrends in Genetics
HOMINID: A framework for identifying associations between host genetic variation and microbiome composition
J. Lynch, K. Tang, S. Priya, J. Sands, M. Sands, E. Tang, S. Mukherjee, D. Knights, and R. Blekhman.GigaScience
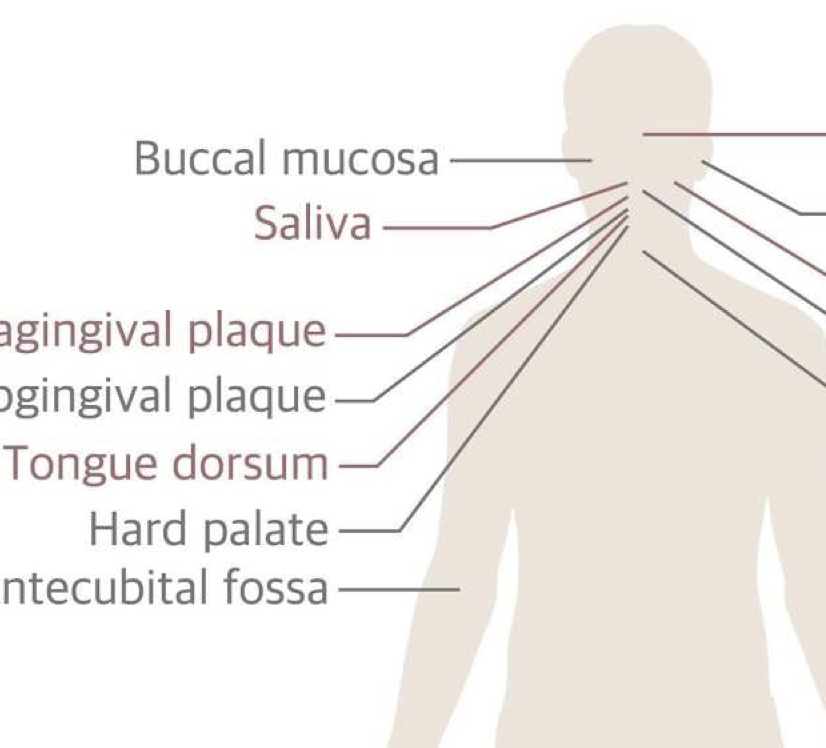
Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation
D. Xu, P. Pavlidis, R. O. Taskent, N. Alachiotis, C. Flanagan, M. DeGiorgio, R. Blekhman, S. Ruhl, and O. GokcumenMolecular Biology and Evolution
Human spit contains ancestral surprises. Science Magazine
Archaic Humans within Spitting Distance, Genetically Speaking. GEN
The secret about human evolution found in spit The Guardian
BugBase predicts organism level microbiome phenotypes
T. Ward, J. Larson, J. Meulemans, B. Hillmann, J. Lynch, D. Sidiropoulos, J. Spear, G. Caporaso, R. Blekhman, R. Knight, R. Fink, and D. Knights.BioRxiv
2016
Gut microbiome of coexisting BaAka pygmies and Bantu reflects gradients of traditional subsistence patterns
A. Gomez, K. Petrzelkova, M. B. Burns, C. J. Yeoman, K. R. Amato, K. Vlckova, D. Modry, A. Todd, C. A. Jost Robinson, M. Remis, M. Torralba, E. Morton, J. D. Umana, F. Carbonero, H. R. Gaskins, K. E. Nelson, B. A. Wilson, R. M. Stumpf, B. A. White, S. R. Leigh, and R. Blekhman.Cell Reports, 2016
Giving Up Hunting and Gathering Changed Our Gut Microbiome. Newsweek
Study Detects Gut Microbiome Differences in African Agriculturist, Hunter-Gatherer Populations. GenomeWeb
The Microbiomes of Indigenous Peoples Are Different--Does It Matter? Scientific American
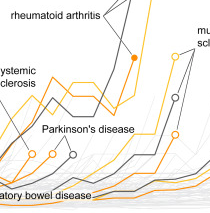
Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens
Y. Nédélec, J. Sanz, G. Baharian, Z. A. Szpiech, A. Pacis, A. Dumaine, J.C. Grenier, A. Freiman, A. J. Sams, S. Hebert, A. P. Sabourin, F. Luca, R. Blekhman, R. D. Hernandez, R. Pique-Regi, J. Tung, V. Yotova, and L. B. Barreiro.Cell, 2016
Neanderthals mated with European humans and it made their immune systems weaker. Quartz
Neanderthal DNA affects ethnic differences in immune response. Nature
Common methods for fecal sample storage in field studies yield consistent signatures of individual identity in microbiome sequencing data
R. Blekhman, K. Tang, E. Archie, L. Barreiro, Z. Johnson, M. Wilson, J. Kohn, M. Yuan, L. Gesquiere, L. Grieneisen, and J. Tung.Scientific Reports, 2016
Atopic Dermatitis Susceptibility Variants In Filaggrin Hitchhike Hornerin Selective Sweep
M. Eaaswarkhanth, D. Xu, C. Flanagan, M. Rzhetskaya, M. G. Hayes, R Blekhman, N. Jablonski, and O. GokcumenGenome Biology and Evolution, 2016
Genetic and transcriptional analysis of human host response to healthy gut microbiome
A. L. Richards, M. B. Burns, A. Alazizi, L. B. Barreiro, R. Pique-Regi, R. Blekhman, and F. Luca.mSystems, 2016
Recent evolution of the salivary mucin MUC7
D. Xu, P. Pavlidis, S. Thamadilok, E. Redwood, S. Fox, R. Blekhman, S. Ruhl, and O. Gokcumen.Scientific Reports, 2016
How your spit Evolved. Popular Science
2015
Host genetic variation impacts microbiome composition across human body sites.
R. Blekhman, J. K. Goodrich, K. Huang, Q. Sun, R. Bukowski, J. T. Bell, T. D. Spector, A. Keinan, R. E. Ley, D. Gevers, and A. G. Clark.Genome Biology, 2015
Genetics, Immunity, and the Microbiome. Karen Zusi, The Scientist
Who is the master of your microbial domain? Jason Tetro, Popular Science
Host genetic architecture and the landscape of microbiome composition: humans weigh in. Andrew Benson, Genome Biology Research Highlight
Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by Entamoeba and Subsistence.
E. R. Morton, J. Lynch, A. Froment, S. Lafosse , E. Heyer , M. Przeworski, R. Blekhman, and L. Segurel.PLOS Genetics, 2015
Surprises Emerge As More Hunter-Gatherer Microbiomes Come In. Ed Yong, National Geographic
Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment.
M. B. Burns, J. Lynch, T. K. Starr, D. Knights, and R. Blekhman.Genome Medicine, 2015
New colon cancer culprit found in gut microbiome. Science Daily
Microbiome Study of Colorectal Cancer Uncovers Signature of Virulence Genes Genome Web
U research furthers options for colorectal treatment and prevention. Minnesota Daily
Linking the microbiome and colorectal cancer: an author Q+A. BioMed Central, On Medicine
Social networks predict gut microbiome composition in wild baboons
J. Tung, L. B. Barreiro, M. B. Burns, J.-C. Grenier, J. Lynch, L. E. Grieneisen, J. Altmann, S. C. Alberts, R. Blekhman, and E. A. Archie.eLife, 2015
Paper (eLife online viewer)
Baboon friends swap gut germs, EurikAlert
Socially structured gut microbiomes in wild baboons, Noah Snyder-Mackler, The Molecular Ecologist
Metagenomics: Social behavior and the microbiome, Jack Gilbert, eLife Insight

Temporal variation selects for diet-microbe co-metabolic traits in the gut of Gorilla spp.
A. Gomez, J. M. Rothman, K. Petrzelkova, C. J. Yeoman, K. Vlckova, J. D. Umana, M. Carr, D. Modry, A. Todd, M. Torralba, K. E. Nelson, R. M. Stumpf, B. A. Wilson, R. Blekhman, B. A. White, and S. R. Leigh.The ISME Journal, 2015
Population genomics analysis of 962 whole genomes of humans reveals natural selection in non-coding regions
F. Yu, J. Lu, X. Liu, E. Gazave, D. Chang, S. Raj, H. Hunter-Zinck, R. Blekhman, L. Arbiza, C. Van Hout, A. Morrison, A. D. Johnson, J. Bis, L. A. Cupples, B. M. Psaty, D. Muzny, J. Yu, R. A. Gibbs, A. Keinan, A. G. Clark, E. Boerwinkle.PLOS One, 2015
2014
Human genetics shape the gut microbiome
J. K. Goodrich, J. L. Waters, A. C. Poole, J. L. Sutter, O. Koren, R. Blekhman, M. Beaumont, W. Van Treuren, R. Knight, J. T. Bell, T. Spector, A. G. Clark, and R. E. Ley.Cell, 2014
Genetics may foster bugs that keep you thin. Science magazine
The Most Heritable Gut Bacterium is… Wait, What is That? National Geographic / Not Exactly Rocket Science
Gut Microbiome Heritability. The Scientist
Body weight heavily influenced by gut microbes: Genes shape body weight by affecting gut microbes. Science Daily
Comparative metabolomics in primates reveals the effects of diet and gene regulatory variation on metabolic divergence
R. Blekhman, G. H. Perry, S. Shahbaz, A. G. Clark, O. Fiehn, and Y. Gilad.Scientific Reports, 2014
Paper (pdf)
2013 and earlier
-
Gene expression differences among primates are associated with changes in a histone epigenetic modification.
C. E. Cain, R. Blekhman, J. C. Marioni, and Y. Gilad.
Genetics, 2011 Apr;184(4)
Paper (pdf)
Appeared in Genetics issue highlights -
Functional comparison of innate immune signaling pathways in primates.
L. B. Barreiro, J. C. Marioni, R. Blekhman, M. Stephens, and Y. Gilad.
PLoS Genetics, 2010 Dec;6(12)
Paper (pdf)
Media coverage: "The Disease Advantage of Chimpanzees", Science Life blog
Media coverage: "Why Humans Are More Sensitive to Certain Viruses", Science Daily
-
Sex-specific and lineage-specific alternative splicing in primates.
R. Blekhman, J. C. Marioni, P. Zumbo, M. Stephens, and Y. Gilad.
Genome Research, 2010 Feb; 20(2): 180-9
Paper (pdf)
Data (zip)
Recommended by Faculty of 1000
Media coverage: "What Makes a Man a Man?", Science Life blog
-
Segmental duplications contribute to gene expression differences between humans and chimpanzees.
R. Blekhman, A. Oshlack, and Y. Gilad.
Genetics, 2009, 182 (627-630)
Paper (pdf)
Appeared in Genetics issue highlights (pdf)
-
A signature of evolutionary constraint on ectopically expressed olfactory receptor genes.
O. De la Cruz, R. Blekhman, X. Zhang, D. Nicolae, S. Firestein, and Y. Gilad.
Molecular Biology and Evolution, 2009 Mar;26(3)
Paper (pdf)
-
Gene regulation in primates evolves under tissue-specific selection pressures.
R. Blekhman, A. Oshlack, A. E. Chabot, G. Smyth, and Y. Gilad.
PLoS Genetics, 2008 Nov;4(11)
Paper (pdf)
Data (zip)
-
Selective constraints in experimentally defined primate regulatory regions.
D. J. Gaffney, R. Blekhman, and J. Majewski.
PLoS Genetics, 2008 Aug 15;4(8)
Paper (pdf)
-
Natural selection on genes that underlie human disease susceptibility.
R. Blekhman, O. Man, L. Herrmann, A. R. Boyko, A. Indap, C. Kosiol, C. D. Bustamante, K. M. Teshima, and M. Przeworski.
Current Biology, 2008 Jun 24;18(12)
Paper (pdf)
Data (tab-delimited text)
Blog coverage: Yann Klimentidis
Blog coverage: Gene Expression
-
An evolutionarily conserved sexual signature in the primate brain.
B. Reinius, P. Saetre, J. A. Leonard, R. Blekhman, R. Merino-Martinez, Y. Gilad, and E. Jazin.
PLoS Genetics, 2008 Jun 20;4(6)
Paper (pdf)
Highlighted in Nature (pdf)
Highlighted in Nature Reviews Neuroscience (pdf)
Media coverage: Times Online
Media coverage: Science Daily
-
A direct transcriptional target of human FOXO1 mediates response to oxidative stress.
P. De Candia, R. Blekhman, A. E. Chabot, A. Oshlack, and Y. Gilad.
PLoS One, 2008, Feb 27;3(2)
Paper (pdf)
-
SPIKE: a database, visualization and analysis tool of cellular signaling pathways.
R. Elkon, R. Vesterman, N. Amit, I. Ulitsky, I. Zohar, M. Weisz, G. Mass, N. Orlev, G. Sternberg, R. Blekhman, J. Assa, Y. Shiloh and R. Shamir.
BMC Bioinformatics, 2008 Feb 20;9:110
Paper (pdf)
SPIKE website
-
Using reporter gene assays to identify cis regulatory differences between humans and chimpanzees.
A. Chabot, R. A. Shrit, R. Blekhman, and Y. Gilad.
Genetics, 2007, 176(4):2069-76
Paper (pdf)
Appeared in Genetics issue highlights
-
The "domino theory" of gene death: gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens.
T. Dagan, R. Blekhman, and D. Graur.
Molecular Biology and Evolution, 2006, 23(2):310-316
Paper (pdf)
Thesis
-
The evolution of gene regulation in primates.
R. Blekhman
Ph.D. thesis, The University of Chicago, 2010