The microbial cells that colonize the human body outnumber host cells by an order of magnitude. Recent advances in high throughput sequencing technologies have unveiled great variability in the microbiome, and shifts in the composition of microbial communities have been associated with multiple health conditions, such as diabetes, inflammatory bowel disease, obesity, and cancer. Although the human microbiome is influenced by environmental factors, microbial composition is also shaped by host genes and genetic variation. In turn, microbes can affect the host by regulating gene expression interacting epithelial cells.
We study host genomic factors that control and interact with the microbiome. We utilize high-throughput genomics technologies and employ computational, statistical, network-theory, data mining, and population genetic analytical approaches, with the goal of understanding how we interact with our microbial communities, how host-microbe interactions affect human disease, and how the symbiosis between us and our microbiome evolved. We aim to answer the following questions:
1. What are the molecular and genetic mechanisms controlling host-microbe interactions?
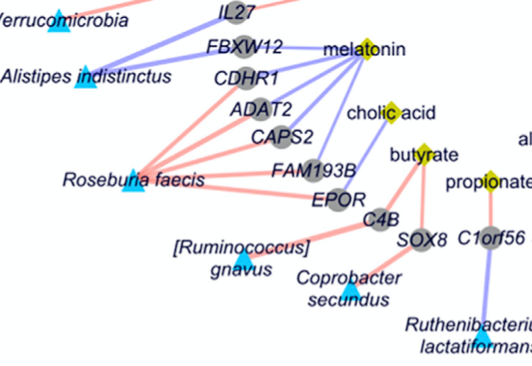
We develop and apply machine learning methods to integrate microbiome and host genomic data to characterize associations between host genes and microbial taxa, and understand how these associations change in health and disease. Recent publications in this area: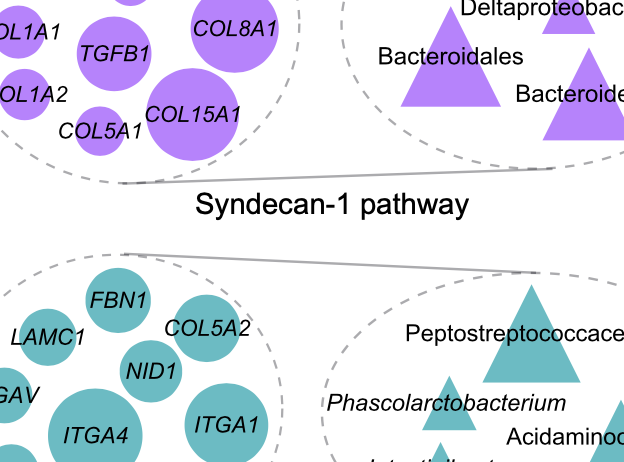
Priya et al., Nature Microbiology, 2022
Mihindukulasuriya et al., Gastroenterology, 2021
Mars et al., Cell, 2020
2. How does host genetic variation impact the microbiome?
What is the heritability of the microbiome, how is it affected by different environmental and host factors, how does it change over time? Recent publications in this area: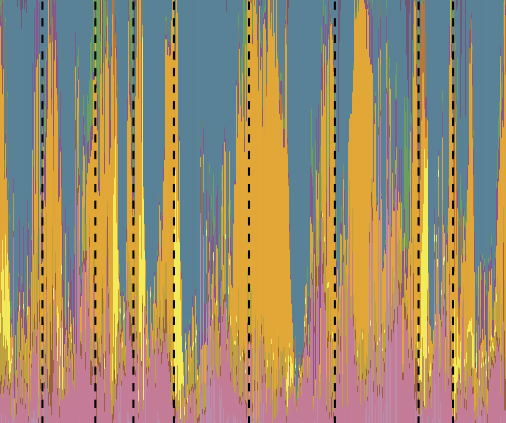
Grieneisen et al., Science, 2021
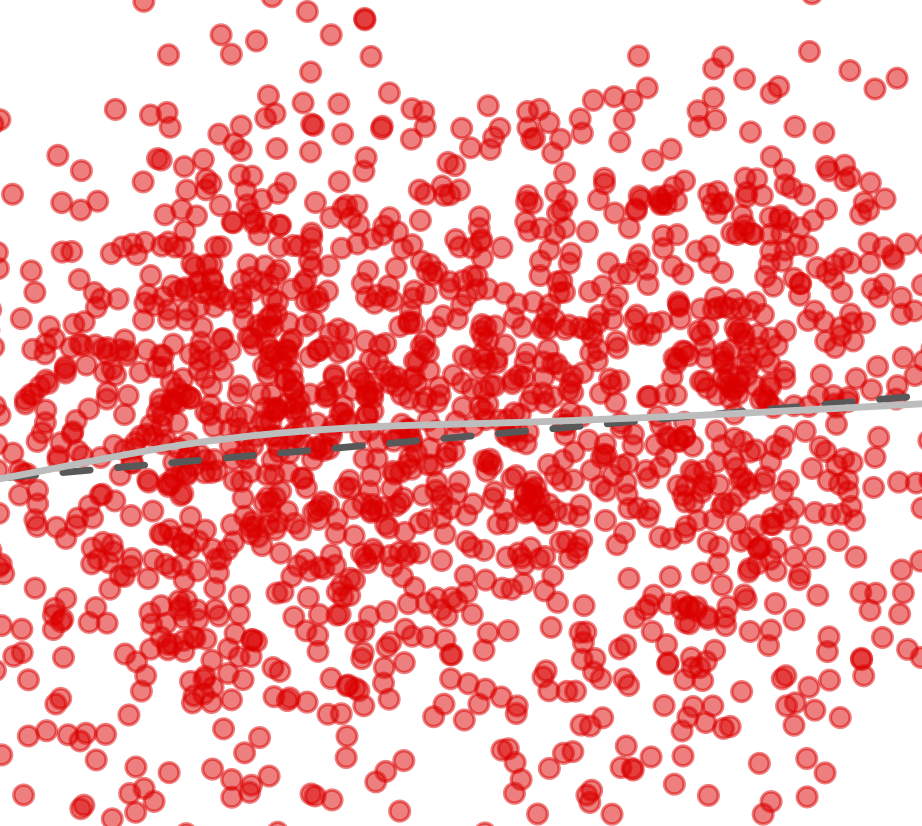
Blekhman et al., Genome Biology, 2015
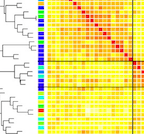
3. What is the effect of variation in the microbiome on host gene regulation?
What are the differences in the microbiome between individuals, human populations, and hominid species, and how do these affect host physiology through regulation of gene expression in interacting host cells? Recent publications in this area: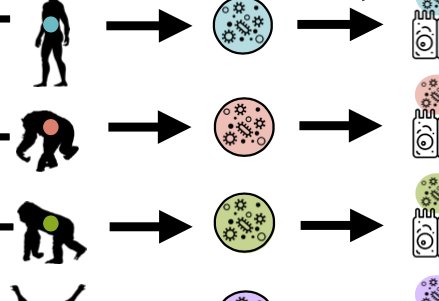
Muehlbauer et al., Cell Reports, 2021
Gomez et al., Cell Reports, 2016
4. How do host-microbiome interactions control susceptibility to colorectal cancer?
What are the unique roles of tumor genetics and microbial communities, and how do they interact to affect cancer susceptibility, progression, and treatment? Recent publications in this area:
Dayama et al., Genome Medicine, 2020
Burns et al., PLOS Genetics, 2018

Yuan et al., mSystems, 2018
5. Meta-research
We mine and analyze large-scale datasets to quantify how research (in microbiome, genomics, and biology) is performed, communicated, and shared, quantify issues and biases, and highlight potential solutions. Recent publications in this area:
Abdill et al., PLOS Biology, 2022


Mangul et al., PLOS Biology, 2019
Collaborators
- Frank Albert (University of Minnesota)
- Beth Archie (University of Notre Dame)
- Luis Barreiro (University of Chicago)
- Eugene Chang (University of Chicago)
- Erika Claud (University of Chicago)
- Sean Davis (University of Colorado)
- Ellen Demerath (University of Minnesota)
- Cheryl Gale (University at Minnesota)
- Andres Gomez (University of Minnesota)
- Casey Greene (University of Colorado)
- Mathieu Groussin (Kiel University)
- Bana Jabri (University of Chicago)
- Dan Knights (University of Minnesota)
- Arjun Krishnan (University of Colorado)
- Sonia Kupfer (University of Chicago)
- Ruth Ley (Max Planck Institute for Biology Tübingen)
- Sam Light (University of Chicago)
- Francesca Luca (University of Chicago)
- Eric Pamer (University of Chicago)
- Roger Pique-Regi (Wayne State University)
- Mathilde Poyet (Kiel University)
- Jenny Tung (Max Planck Institute for Evolutionary Anthropology)
Funding
We are thankful to the various funding agencies and institutions that have supported our research throughout the years, including the National Institutes of Health, National Science Foundation, Alfred P. Sloan Foundation, American Cancer Society, University of Minnesota, and University of Chicago.
Our lab is currently funded through the following grant awards:
- NIH/NIGMS R35-GM128716: Population Genomics of Host-Microbiome Interactions
- NIH/NICHD R01-HD109830: Milk-Omics: Systems Biology of Human Milk and Its Links to Maternal and Infant Health
- NIH/NLM R01-LM013863: Human Microbiome Compendium: large-scale curation and processing of human microbiome datasets
- NIH/NIDDK P30-DK042086: Center for Interdisciplinary Study of Inflammatory Intestinal Disorders