Our Research
We study how human genes can control and interact with the microbiome. We utilize high-throughput genomics technologies and employ computational, statistical, machine learning, and population genetic analytical approaches, with the goal of understanding how we interact with our microbial communities, how host-microbe interactions affect human disease, and how the symbiosis between us and our microbiome evolved.
- Read more about our research and latest publications
- See our new lab location on the University of Chicago campus
- We're hiring! See how you can join the lab

Key Recent Publications
Integration of 168,000 samples reveals global patterns of the human gut microbiome
Abdill, Graham, et al., Cell, 2025
Abdill, Graham, et al., Cell, 2025
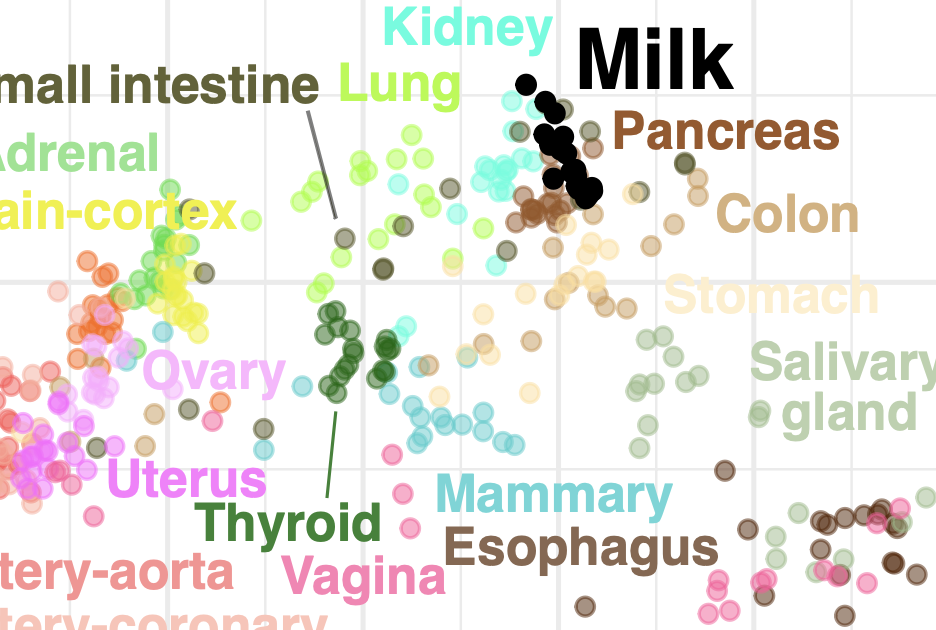
Human milk variation is shaped by maternal genetics and impacts the infant gut microbiome
Johnson et al., Cell Genomics, 2024
Johnson et al., Cell Genomics, 2024
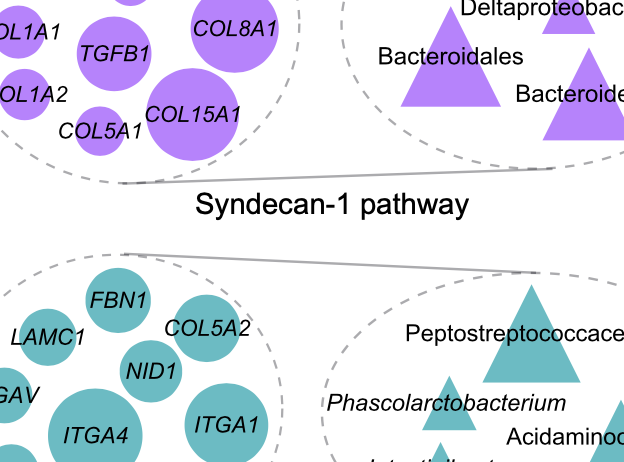
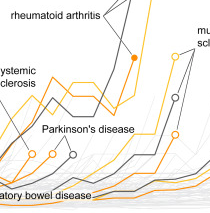
Identification of shared and disease-specific host gene–microbiome associations across human diseases using multi-omic integration
Priya et al., Nature Microbiology, 2022
Priya et al., Nature Microbiology, 2022
Public human microbiome data are dominated by highly developed countries
Abdill et al., PLOS Biology, 2022
Abdill et al., PLOS Biology, 2022
Gut microbiome heritability is nearly universal but environmentally contingent
Grieneisen et al., Science, 2021
Grieneisen et al., Science, 2021
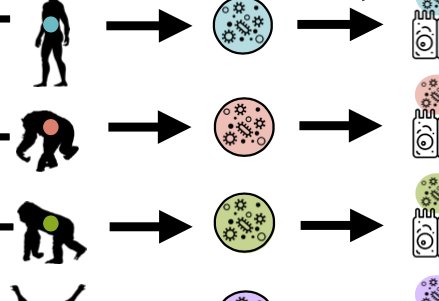
Interspecies variation in hominid gut microbiota controls host gene regulation
Muehlbauer et al., Cell Reports, 2021
Muehlbauer et al., Cell Reports, 2021
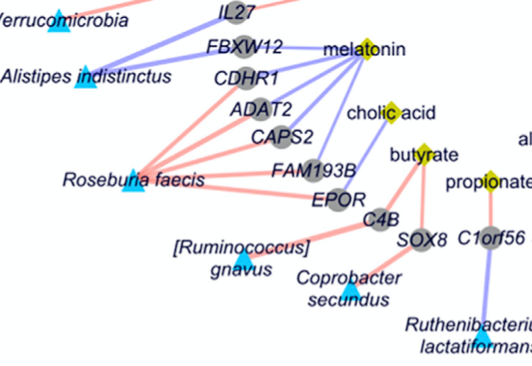
Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome
Mars et al., Cell, 2020
Mars et al., Cell, 2020
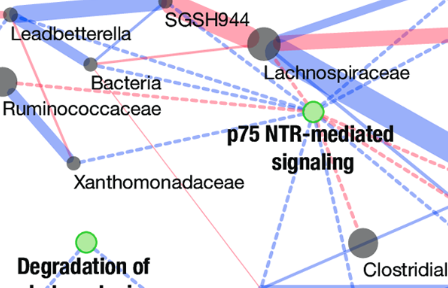
Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment
Burns et al., PLOS Genetics, 2018
Burns et al., PLOS Genetics, 2018
Genetic ancestry and natural selection drive population differences in immune responses to pathogens
Nédélec et al. Cell, 2016
Nédélec et al. Cell, 2016
Gut microbiome of coexisting BaAka pygmies and Bantu reflects gradients of traditional subsistence patterns
Gomez et al., Cell Reports, 2016
Gomez et al., Cell Reports, 2016
Host genetic variation impacts microbiome composition across human body sites
Blekhman et al., Genome Biology, 2015
Blekhman et al., Genome Biology, 2015
Lab News
- Welcome to new MD-PhD student Tess Brunner! 10/2024
- Congrats to Dr. Sambhawa Priya on being named a Chicago DFI Fellow! 09/2024
- Congrats to Dr. Mattea Allert on defending her PhD! 08/2024
- Congrats to Dr. Kelsey Johnson on her NIH K99 award! 07/2024
- Welcome to new postdoc Dr. Sambhawa Priya! 06/2024
- Welcome to new graduate student Liz Gibbons! 08/2023
- Congrats to Dr. Mark Swanson for being awarded an NSF Postdoctoral Fellowship in Biology! 07/2023
- Welcome to new lab members JJ Colgan and Marco Rossi! 04/2023
- Welcome to new lab members Rich Abdill and Pamela Ferretti! 01/2023
- Welcome to new lab member Katja Della Libera! 09/2022
- Our lab has moved to the University of Chicago! And we're hiring at all levels -- see how you can join the lab! 08/2022
- Congrats to Dr. Amanda Muehlbauer for successfully defending her PhD! 07/2022
- Congrats to Dr. Laura Grieneisen on her new faculty position at The University of British Columbia! 05/2022
- Congrats to Dr. Rich Abdill for successfully defending his PhD! 02/2022
- Congrats to Dr. Sambhawa Priya for successfully defending her PhD! 10/2021
- Welcome to new lab member Ashwin Chetty! 09/2021
- Congrats to Sabrina Arif for being selected to the Genetics and Genomics Predoctoral Training Grant ! 07/2021
- Congratulations to Mattea Allert on receiving the President’s Student Leadership and Service Award! 05/2021
- Congrats to Dr. Kelsey Johnson on her NIH NRSA postdoctoral fellowship! 04/2021
- Welcome to new lab members Sabrina Arif and Samantha Graham! 04/2021
- Congrats to Dr. Beth Adamowicz for being awarded a Minnesota Colorectal Cancer Research Foundation Grant! 02/2021
- We have been awarded a Masonic Cross-Departmental Grant in Children’s Health to study host-microbiome interactions in human milk 01/2021
- Congrats to Dr. Beth Adamowicz for winning the UMN CBS Postdoc Award for Outreach and Communication! 09/2020
- Congrats to Dr. Kelsey Johnson for being selected as a MinnCResT NIH T32 Fellow! 06/2020
- Welcome to Kelsey Johnson, new postdoctoral associate in the lab! 01/2020
- Welcome to new postdoctoral associate Beth Adamowicz! 07/2019
- Congrats to Rich Abdill for receiving the best talk award at the 2019 Great Lakes Bioinformatics Conference 05/2019
- Congrats to Sambhawa Priya for being awarded the UMN Doctoral Dissertation Fellowship! 04/2019
- Sambhawa Priya will speak at the Workshop on Statistical and Algorithmic Challenges in Microbiome Data Analysis in NYC 03/2019
- Congrats to Laura Grieneisen for winning the best postdoc talk award at the 2018 Midwest Popgen Meeting! 08/2018
- Welcome to new lab members Mattea Allert and Rich Abdill! 08/2018
- Our lab has been awarded a MIRA grant from the NIH to support our research for the next five years 07/2018
- Congrats to Amanda Muehlbauer for being awarded the 2018 Ray Anderson Zoology and Genetics Fellowship! 04/2018
- Ran was named 2018-20 McKnight Land-Grant Professor 02/2018
- Congrats to Sambhawa Priya for winning the best poster award in the Bioinformatics and Computational Biology Research Symposium! 01/2018
- We have been awarded a grant from the Minnesota Partnership for Biotechnology and Medical Genomics to study the role of the gut microbiome in colorectal cancer 01/2018
- Congrats to Andres Gomez for his new position as Assistant Professor here at the University of Minnesota! 08/2017
- Awesome work by Amanda, Gargi, and Sambhawa sharing microbiome science with hands-on activities at the Farmers Market! 06/2017
- Ran will speak at the 2017 Biology of Genomes Meeting 05/2017
- Congrats to Laura Grieneisen for being awarded the Grand Challenges in Biology Postdoctoral Fellowship! 04/2017
- Ran will speak at the American Association of Physical Anthropologists (AAPA) Annual Meeting 04/2017
- Welcome to Trevor Gould, a new graduate student in the lab! 04/2017
- Ran will speak at the Plant and Animal Genome XXV Conference 01/2017
- Welcome to new lab members Gargi Dayama, Sambhawa Pryia, and Amanda Muehlbauer! 08/2016
- Ran will speak at the 2016 META Center Symposium on Host-Microbe Systems Biology 07/2016
- Congrats to Michael Burns for his new position as Assistant Professor at Loyola University starting this summer! 06/2016
- Ran will speak at the 2016 Biology of Genomes Meeting 05/2016
- Ran is selected as an Alfred P. Sloan Research Fellow (PDF of New York Times announcement) 02/2016
- We have been awarded a grant from the Randy Shaver Cancer Research Fund to support our research on host-microbiome interaction in colon cancer 01/2016
- Ran will speak at the ASHG Annual Meeting on October 9, 2015 10/2015
- Michael wins the Best Poster award at the UMN 24th Annual Developmental Biology Symposium on Symbiosis and Development. Congrats Michael! 09/2015
- Michael has been awarded a UMN CTSI Translational Research Development Program Grant. Congrats Michael! 05/2015
- Josh has successfully defended his M.S. thesis. Congrats Josh! 08/2015
- Ran will speak at the Charles Schachtele Memorial Symposium in Oral Biology on May 19, 2015 05/2015
- Elise will speak at the UMN Microbiome Symposium on April 17, 2015 04/2015
- Ran will speak at the Gordon Research Conference for Quantitative Genetics and Genomics on February 27, 2015 02/2015
- Josh has been awarded a BICB fellowship for 2015. Congrats Josh! 11/2014
- Tim receives a UROP award for his undergraduate research project. Congrats Tim! 11/2014
- Josh wins second-best poster in the BICB symposium. Congrats Josh! 08/2014
- Andres Gomez joins the lab as a Postdoctoral Associate. Welcome Andres! 06/2014
- Ran will give a talk at the 2014 ASM meeting on May 20 at 3:00 PM, as part of the symposium on Host Genetic Control of the Microbiome, in room 160A at the Boston Convention and Exhibition Center 05/2014
- Michael will present his research at the Biology of Genomes Meeting at Cold Spring Harbor in the poster session on Wednesday May 7, 2014 05/2014
- Ran will give a talk at the AAPA Meeting on April 12, 2014 at 11:15 AM, as part of the Beyond The Genome Symposium, at the Hyatt Imperial Ballroom, TELUS Convention Center, Calgary, Canada 04/2014
- Elise Morton joins the lab as a Postdoctoral Associate. Welcome Elise! 02/2014
- Josh Lynch joins the lab as a BICB graduate student. Welcome Josh! 01/2014
- Karen Tang joins the lab as a data analyst. Welcome Karen! 01/2014
- Ran will give a talk at the ASHG Meeting on October 24, 2013 at 2:00 PM, at the Grand Ballroom East, Level 3 of the Boston Convention & Exhibition Center 10/2013
- Ran will give a talk at the NTD Conference in Austin on October 9, 2013 09/2013
- Ran will give a talk at the GCD Seminar Series on September 19, 2013 09/2013
- Michael Burns joins the lab as an HHMI Postdoctoral Associate. Welcome Michael! 08/2013
- Ran will give a talk at the MPGI Fall Symposium at the University of Minnesota on August 29, 2013 08/2013
- The Blekhman Lab opened its gates on August 1st, 2013 in the Cargill Building on the St. Paul campus! 08/2013
- Ran will give a talk at the SMBE Meeting in Chicago on July 10, 2013 06/2013
- Ran will give a talk at the Biology of Genomes meeting at Cold Spring Harbor on May 8, 2013 05/2013
The Human Microbiome Compendium: An ongoing project to build a dataset of the human microbiome at an unprecedented scale
Integration of 168,000 samples reveals global patterns of the human gut microbiome (Cell, 2025)Full dataset for download, version 1.1 (Zenodo)
MicroBioMap R package (github)
Website (microbiomap.org)
An R package for visualizing microbiome time series data
R package, code, and user guide (GitHub)Publication (mSystems, 2022)
A framework for identifying host genetic variants associated with microbiome composition
Software, code, and instructions (GitHub)Online network visualization tool
Paper (GigaScience)
Our Work in the Media
-

The Human Microbiome Compendium on the cover of Cell
Cell, 2025